By Jamie Southgate, Rangarirai Matima and Tracey-Calvert Joshua
Introduction
2023 was a great year for the PHA4GE consortium as it hosted its inaugural conference in Cape Town, South Africa. Interest in joining the various Working Groups rose as individuals heard about the various projects being undertaken, technical expertise shared and camaraderie in handling common global challenges.
As PHA4GE has experienced immense growth in the past year, the Secretariat proposed the idea of exploring the interests and geographical diversity of PHA4GE membership as well as the proficiency of skills within our community, to effectively prepare and guide PHA4GE in undertaking future projects and activities.
In April and June 2024, we invited all PHA4GE members (n=424) to take part in a survey. Respondents included a global coalition of individuals actively working in PHA4GE Working Groups (n=347) and other individuals interested in receiving updates on the work we do (n=77). The six PHA4GE Working Groups (WGs) which focus on microbial genomics, include: Bioinformatics Pipelines and Data Visualization, Data Repositories, Data Structures, Ethics and Data Sharing, Infrastructure and Training and Workforce Development. These members range from early career scientists (i.e., students) to seasoned experts in the field (i.e., distinguished researchers and academia, policy makers, senior public health labs and public healthcare officials).
A huge thank you to all who participated in the survey and we hope that sharing our insights will help you navigate the shifting genomics landscape with greater confidence.
Survey Results
Affiliation by Country/Region

The diverse representation of countries/regions amongst PHA4GE members continues to expand and we received responses from 30 out of 60 listed member countries as shown in Figure 1.
The highest number of respondents were from the United States (15.5%) and South Africa (9.9%), followed by the United Kingdom (8.5%). It was encouraging to see that a large proportion (55.9 %) of the respondents were located in low-to-middle income countries (LMICs), which indicates a broad international reach and interest in PHA4GE activities.
Membership in PHA4GE Working Groups
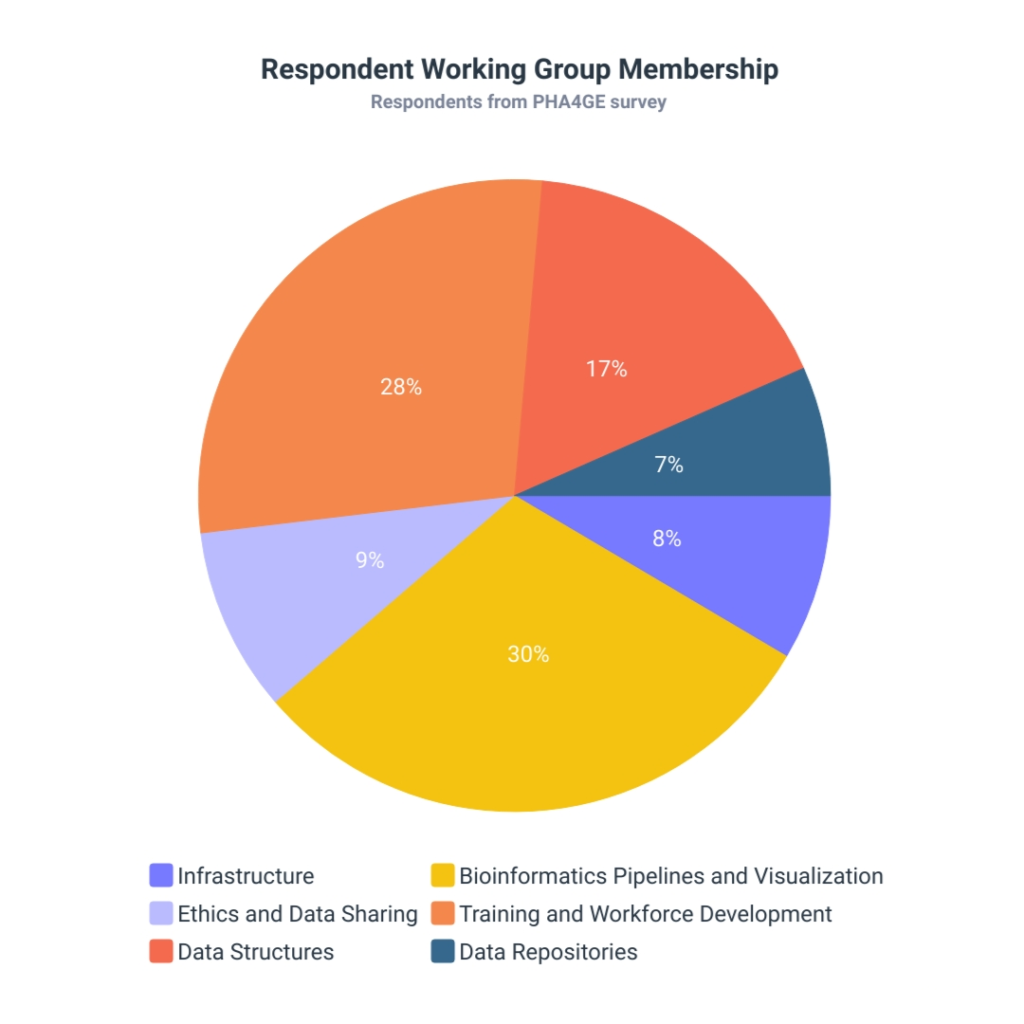
All respondents were members of the PHA4GE Working Groups (n=72) – which translates to a 20.7% response rate. The Working Group with the largest response rate (30%) was the Bioinformatics Pipelines and Visualization Working Group; which concurrently has the largest number of members in the consortium (Figure 2).
Infectious Disease interest
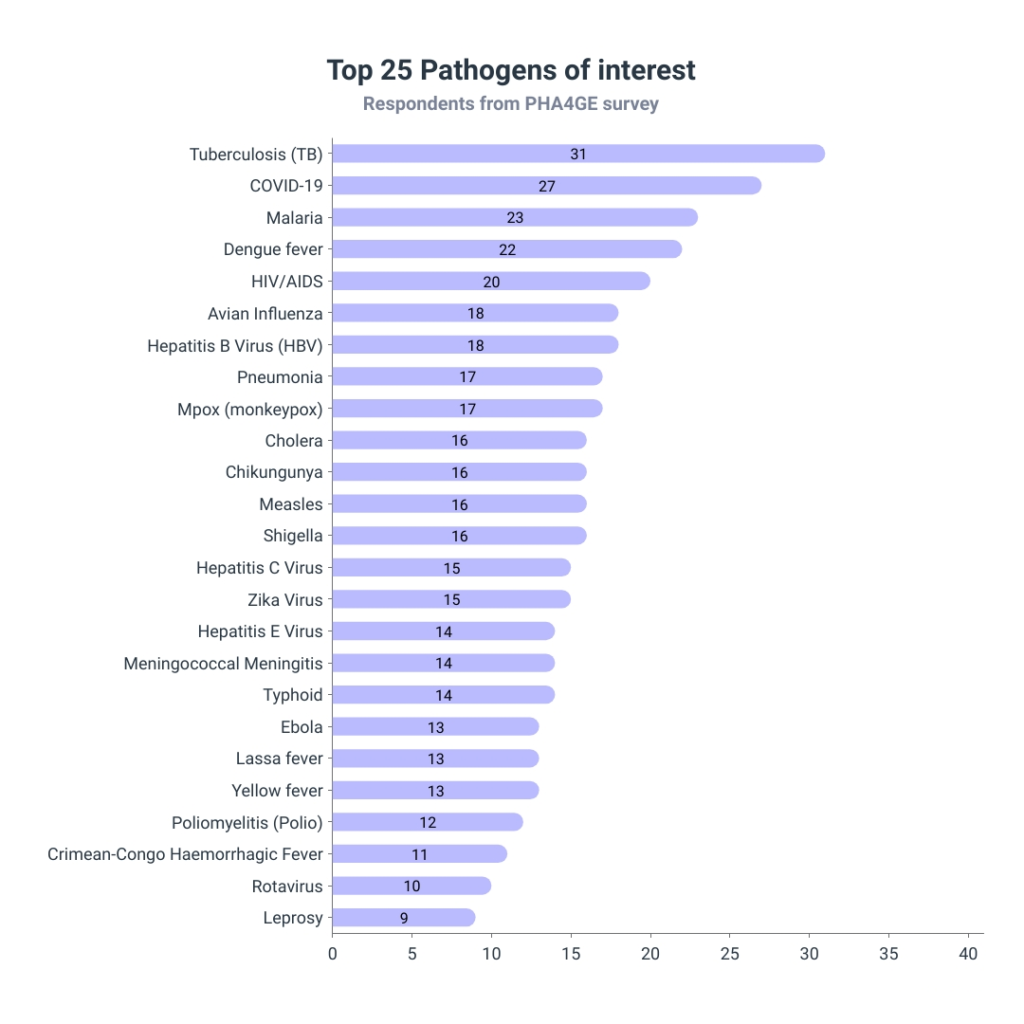
Participants were asked to select their diseases of interest out of a list of (n = 39) current global diseases of interest. As shown in Figure 3, the two most fatal infectious diseases, tuberculosis and COVID-19 (Falzon et al., 2023), were also the most frequently selected. This was followed by Malaria, Dengue fever and HIV/AIDS. Tuberculosis, HIV/AIDS and Malaria are known as the Big Three, due to the immense number of infections and fatalities they cause each year (Makam and Matsa, 2021). Dengue fever remains endemic in over 100 countries (Africa, the Americas, the Eastern Mediterranean, South-East Asia and the Western Pacific), threatening the health and well-being of nearly half of the world’s population, with a projection of nearly 3.9 billion people being a risk (WHO, 2024).
An additional 12 responses were given for “other” diseases of interest. Some of which were “common” diseases, categories of broad microbes, preventive methods or interventions that may be applied in microbial genomics, and non-microbial genomics; more specifically, diseases related to neurodivergent conditions (Table 1)
| Additional Interests | Number of respondents |
| Common Diseases | |
| Influenza | 1 |
| Diarrhoeal diseases | 1 |
| Categories of broad microbes | |
| Respiratory syncytial virus (RSV) | 1 |
| MetaPneumovirus | 1 |
| Campylobacter | 1 |
| Salmonella | 1 |
| Any opportunistic bacterial pathogens | 2 |
| Shiga toxin E. coli | 1 |
| Preventive methods or interventions that may be applied in microbial genomics | |
| Ethics and Data Sharing | 1 |
| Antimicrobial Resistance (AMR) | 4 |
| Non-microbial genomics | |
| Care for Children and Adolescents on the Spectrum and other neurodivergent individuals who present an atypical neurocognitive condition | 1 |
Summary of Experience Levels
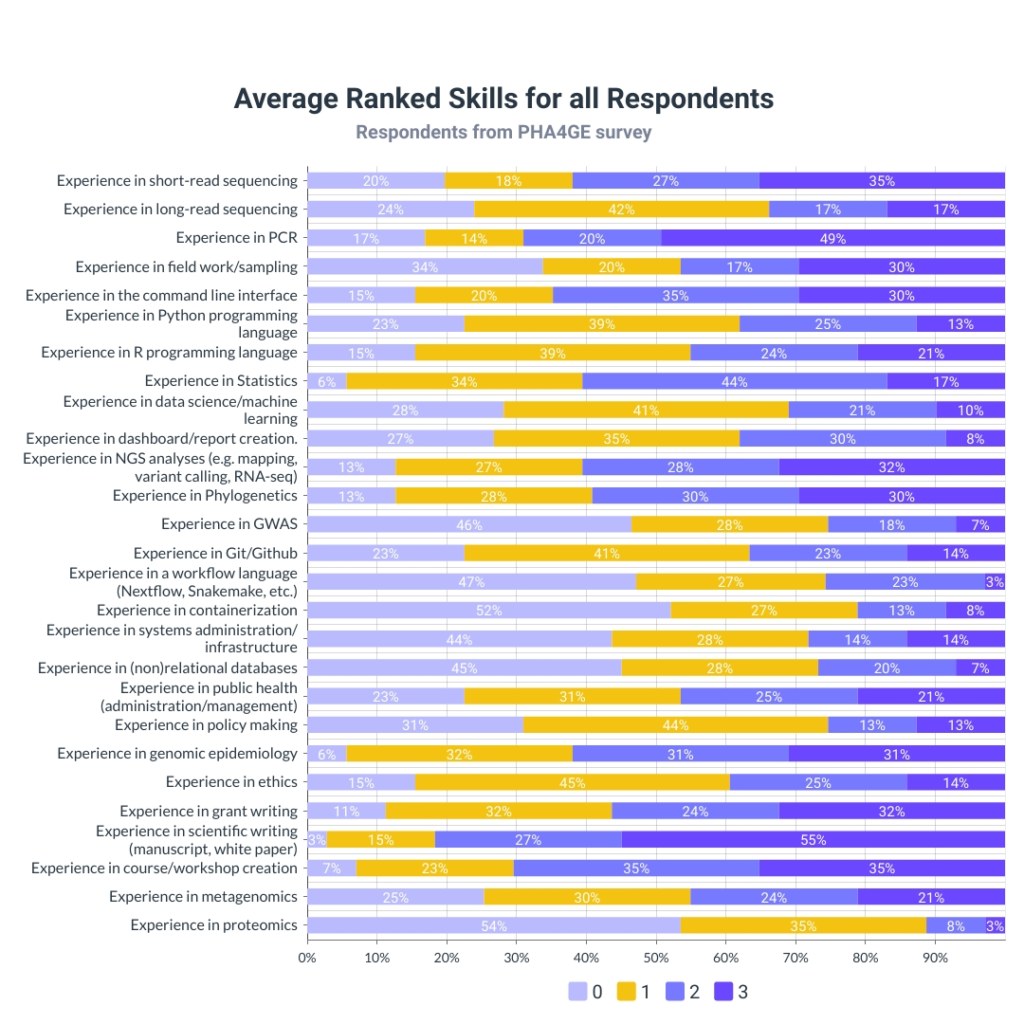
Further questions were centered around wet and dry lab skills, systems administration, scientific writing and reporting. Additional skills relating to public health practitioners, covered knowledge on policy making, training, sourcing funding for projects (grant writing) and ethics.
Figure 4 shows the average ranking of all skills. A Likert scale was used, ranging from 0 to 3; where 0 was least skilled, and 3 was at expert level. We calculated the average for each skill to determine the representative percentage of the strength of each skill from all the respondents.
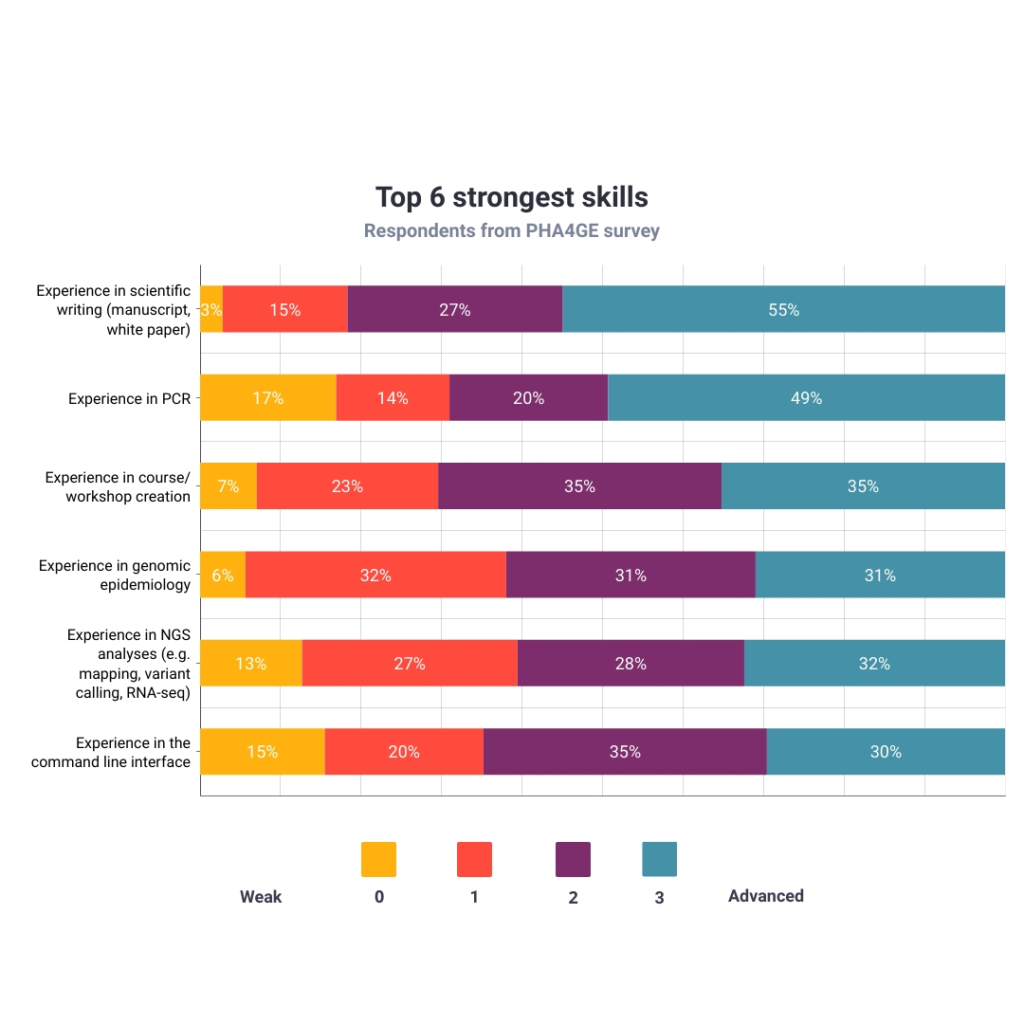
Figure 5 shows the highly-ranked skills. The top three strongest skills were: experience in scientific writing (55%), experience in running polymerase chain reaction (PCR) tests (49%) and experience in creating content and running courses/workshops (35%).
Since the vision of PHA4GE is to strengthen public health bioinformatics and epidemiology, it was not surprising that NGS analyses (32%), genomic epidemiology (31%) and working with the command line interface (30%) were some of the most honed skills.
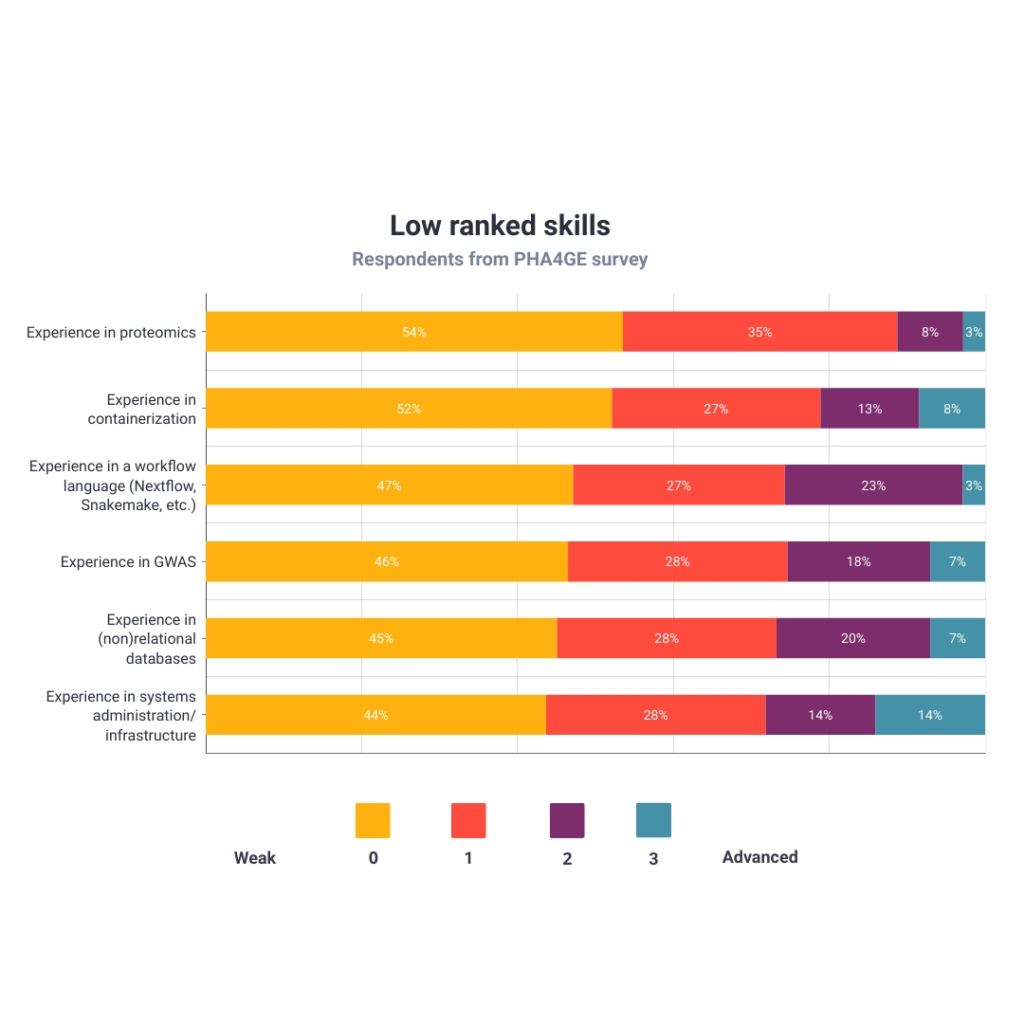
Figure 6 shows the six most underdeveloped skills. The three weakest skills were experience in proteomics (54%), experience in containerization (52%) and experience in Workflow languages (47%). Notably, most of these are related to more technical work such as computing infrastructure and advanced levels of coding. The Infrastructure WG, where many of these skills are expected to be utilized, was one of the smallest groups of respondents.
These insights provide a comprehensive understanding of the respondent demographics, their involvement in PHA4GE, and their areas of expertise. This information can be used to tailor future projects, events, and communications to better meet the needs and interests of PHA4GE members.
Key take-aways from the results
- The results highlight a gap in computational skills related to computational infrastructure that are increasingly necessary for managing and analyzing large-scale biological data sets.
- In 2016 Helmy et al., showed that challenges in bioinformatics capacity were largely attributed to lack of resources in developing countries. However, efforts in bioinformatics capacity are evident (Mboowa et al., 2021) as was shown by NGS analyses being one of the strongest skills across the globally-represented respondents.
- The pathogens of interest indicate the priority public health needs globally. The “Other interests” category showed the preference of scientists to work on pathogens more broadly, rather than being restricted to one disease of interest (Kwok et al., 2021).
Limitations
- A low response rate may have contributed to a bias in the results.
- The surveyed skills are not exhaustive of dry lab and wet lab skills required in bioinformatics
- The preferred pathogens of interest may be skewed towards the usually highly-funded pathogens
Conclusion
The findings from this survey provide insights into the skill distribution and diseases of interests within the PHA4GE community. These results can be utilized to enhance project team compositions by identifying individuals with complementary skills and shared interests in specific pathogens.
Leveraging these findings to build networks based on shared interests can foster collaborative opportunities and facilitate knowledge exchange across different regions.
References
Falzon D, Zignol M, Bastard M, Floyd K, Kasaeva T. The impact of the COVID-19 pandemic on the global tuberculosis epidemic. Front Immunol. 2023 Aug 29;14:1234785. doi: 10.3389/fimmu.2023.1234785. PMID: 37795102; PMCID: PMC10546619.
Helmy, M., Awad, M. and Mosa, K.A., 2016. Limited resources of genome sequencing in developing countries: challenges and solutions. Applied & translational genomics, 9, pp.15-19.
Kwok, A.J., Mentzer, A. and Knight, J.C., 2021. Host genetics and infectious disease: new tools, insights and translational opportunities. Nature Reviews Genetics, 22(3), pp.137-153.
Makam P, Matsa R. “Big Three” Infectious Diseases: Tuberculosis, Malaria and HIV/AIDS. Curr Top Med Chem. 2021;21(31):2779-2799. doi: 10.2174/1568026621666210916170417. PMID: 34530712.
Mboowa, G., Sserwadda, I. and Aruhomukama, D., 2021. Genomics and bioinformatics capacity in Africa: no continent is left behind. Genome, 64(5), pp.503-513.
World Health Organization (WHO). Dengue and severe dengue (2024). https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue. Accessed 31 July 2024



